Projects
Rank-INR
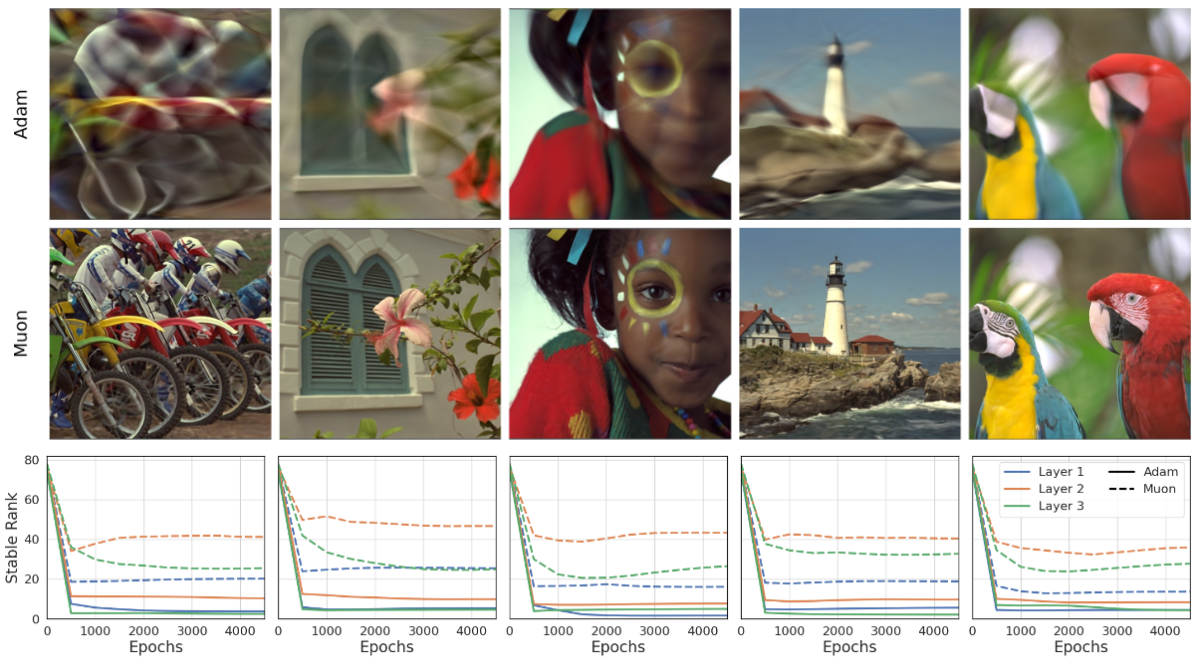
Implicit Neural Representations (INRs) based on vanilla Multi-Layer Perceptrons (MLPs) are widely believed to be incapable of representing high-frequency content. This has directed research efforts towards architectural interventions, such as coordinate embeddings or specialized activation functions. Here, we challenge the notion that the low-frequency bias of vanilla MLPs is an intrinsic, architectural limitation, but instead a symptom of stable rank degradation during training. We empirically demonstrate that regulating the network’s rank during training substantially improves the fidelity of the learned signal, rendering even simple MLP architectures expressive.
| [Paper] | [Code] |
Lipschitz-INR
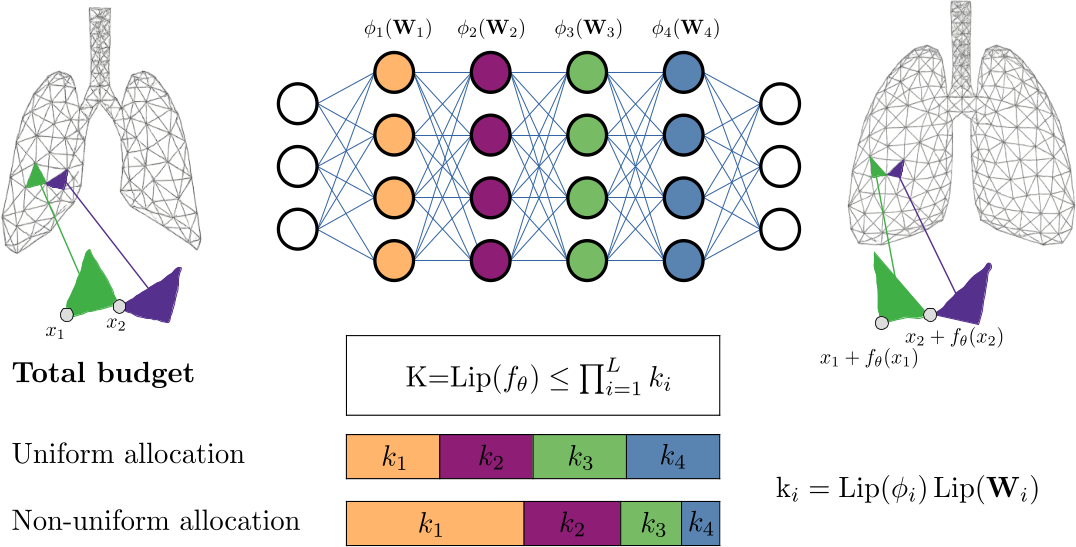
Implicit Neural Representations (INRs) have shown great promise in solving inverse problems, but their lack of inherent regularization often leads to a trade-off between expressiveness and smoothness. While Lipschitz continuity presents a principled form of implicit regularization, it is often applied as a rigid, uniform constraint. In this work, we reframe Lipschitz regularization as a flexible Lipschitz budget framework. We propose a method to first derive a principled, task-specific total budget $K$, then distribute it non-uniformly across all network components, including linear weights, activations, and embeddings. Across extensive experiments on deformable registration and image inpainting, we show that non-uniform allocation strategies provide a measure to balance regularization and expressiveness within the specified global budget.
| [Paper] | [Code] |
MM-DINOv2
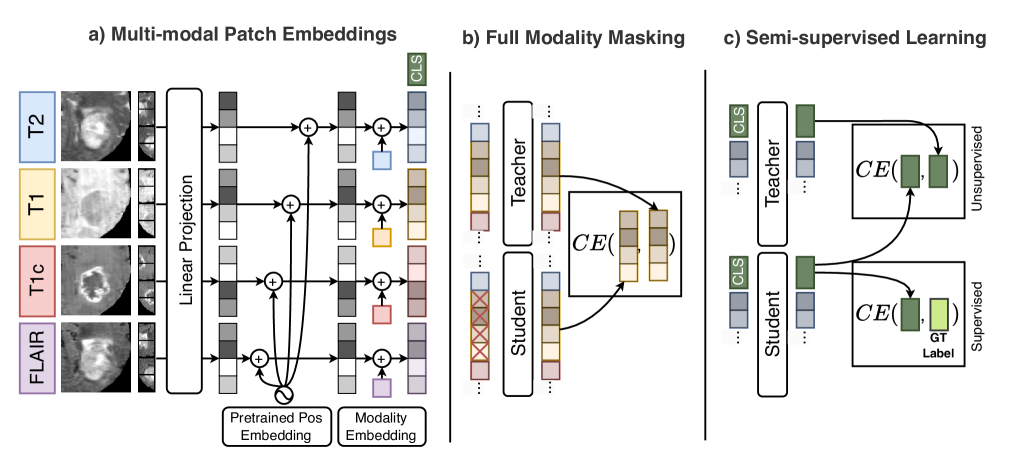
Vision foundation models (VFMs) pre-trained on natural images, such as DINOv2, are powerful yet inherently limited to unimodal analysis, restricting their use in multimodal medical imaging. Supervised baselines handle multi-modal inputs but underuse unlabeled data and degrade when modalities are missing. We propose MM-DINOv2, an efficient adaptation of DINOv2 for multi-modal medical imaging. MM-DINOv2 introduces multi-modal patch embeddings to jointly process heterogeneous inputs, full-modality masking to learn robust cross-modality representations under missing data, and semi-supervised learning to exploit large unlabeled datasets. MM-DINOv2 is a scalable framework that leverages natural-image pretraining while addressing practical clinical challenges of missing data and limited annotations.
| [Paper] | [Code] |
DeepISLES
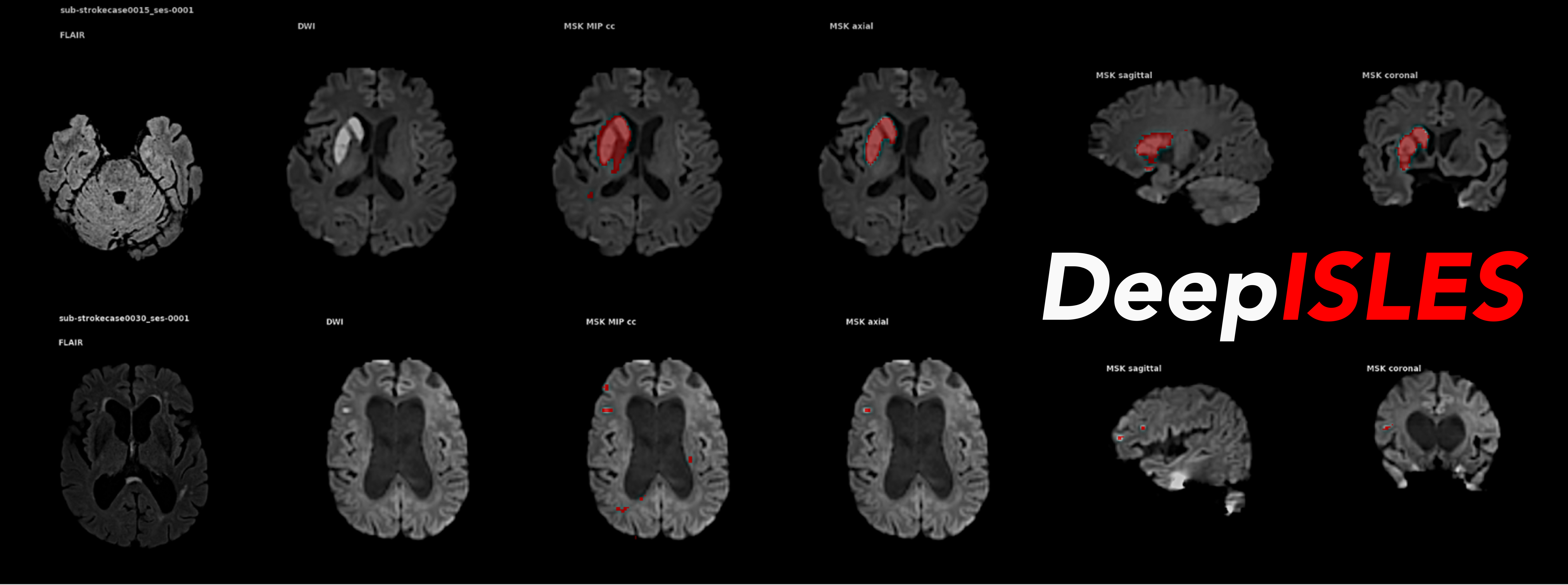
DeepISLES, an AI segmentation ensemble for stroke MRI originating from the ISLES’22 challenge, outperforms previous stroke segmentation models by > 10% and matches expert radiologist performance in identifying stroke lesions, showing strong clinical relevance and generalizability. DeepISLES further highlights the potential of biomedical challenges to bring together leading research groups to jointly advance the field.
| [Paper] | [Code] |
NOVA
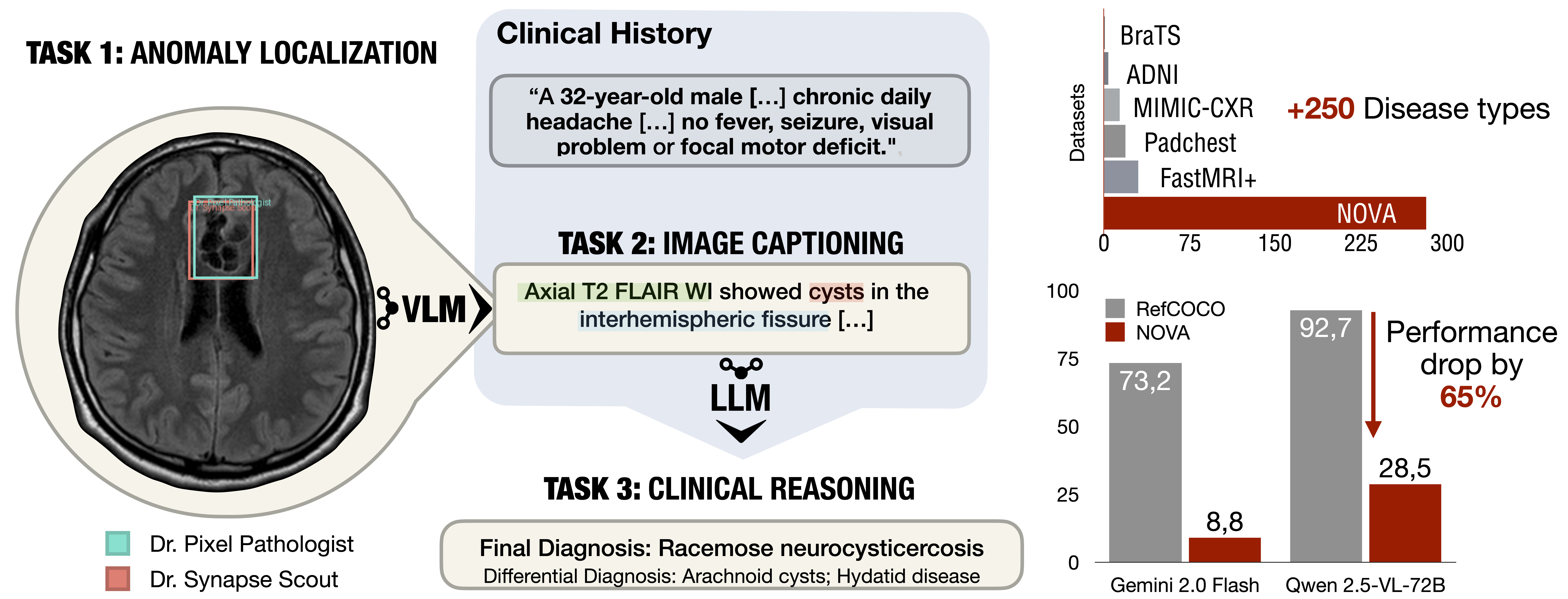 Deployed models often encounter data distributions that differ from their training distributions, requiring robust out-of-distribution detection and open-world recognition. While foundation and vision-language models are expected to generalize broadly, current benchmarks with limited outlier types fail to capture real-world challenges, particularly in clinical settings. We present NOVA, a challenging evaluation benchmark of ~900 brain MRI scans spanning 281 rare pathologies with heterogeneous acquisition protocols. Each case includes clinical narratives and expert bounding-box annotations, enabling joint assessment of anomaly localization, visual captioning, and diagnostic reasoning. As a training-free benchmark, NOVA provides an extreme stress-test of out-of-distribution generalization across both sample appearance and semantic domains, addressing critical gaps in evaluating model robustness for real-world medical applications.
Deployed models often encounter data distributions that differ from their training distributions, requiring robust out-of-distribution detection and open-world recognition. While foundation and vision-language models are expected to generalize broadly, current benchmarks with limited outlier types fail to capture real-world challenges, particularly in clinical settings. We present NOVA, a challenging evaluation benchmark of ~900 brain MRI scans spanning 281 rare pathologies with heterogeneous acquisition protocols. Each case includes clinical narratives and expert bounding-box annotations, enabling joint assessment of anomaly localization, visual captioning, and diagnostic reasoning. As a training-free benchmark, NOVA provides an extreme stress-test of out-of-distribution generalization across both sample appearance and semantic domains, addressing critical gaps in evaluating model robustness for real-world medical applications.
| [Paper] | [Data] |
GliODIL
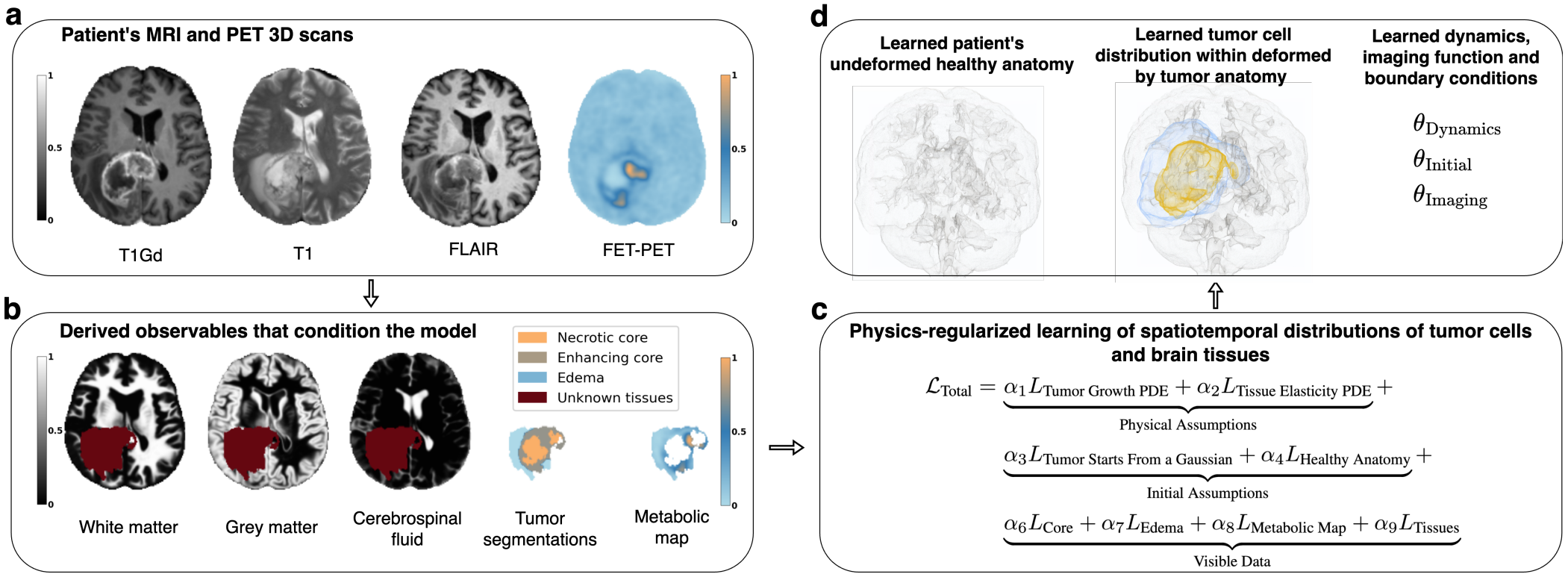 Physical models, represented by partial differential equations, are crucial in addressing many under-constrained problems, such as tumor growth modeling. Deep learning methods often struggle to accurately estimate the full distribution of tumor cells, primarily because of insufficient training data. As a result, most existing approaches rely on physics-based simulations to align with observed tumor characteristics, yielding anatomically and physiologically plausible estimates. However, these methods face challenges when dealing with complex and unknown initial conditions and are often constrained by rigid physical models. In this work, we introduce a novel method that balances data-driven and physics-based cost functions. Specifically, we propose a unique discretization scheme that quantifies how well our learned spatiotemporal distributions of tumors and brain tissue adhere to their respective growth and elasticity equations. This quantification serves as a regularization term rather than a strict constraint, enabling greater flexibility and more effective integration of patient data than existing models.
Physical models, represented by partial differential equations, are crucial in addressing many under-constrained problems, such as tumor growth modeling. Deep learning methods often struggle to accurately estimate the full distribution of tumor cells, primarily because of insufficient training data. As a result, most existing approaches rely on physics-based simulations to align with observed tumor characteristics, yielding anatomically and physiologically plausible estimates. However, these methods face challenges when dealing with complex and unknown initial conditions and are often constrained by rigid physical models. In this work, we introduce a novel method that balances data-driven and physics-based cost functions. Specifically, we propose a unique discretization scheme that quantifies how well our learned spatiotemporal distributions of tumors and brain tissue adhere to their respective growth and elasticity equations. This quantification serves as a regularization term rather than a strict constraint, enabling greater flexibility and more effective integration of patient data than existing models.
| [Paper] | [Code] |
DL-Prior
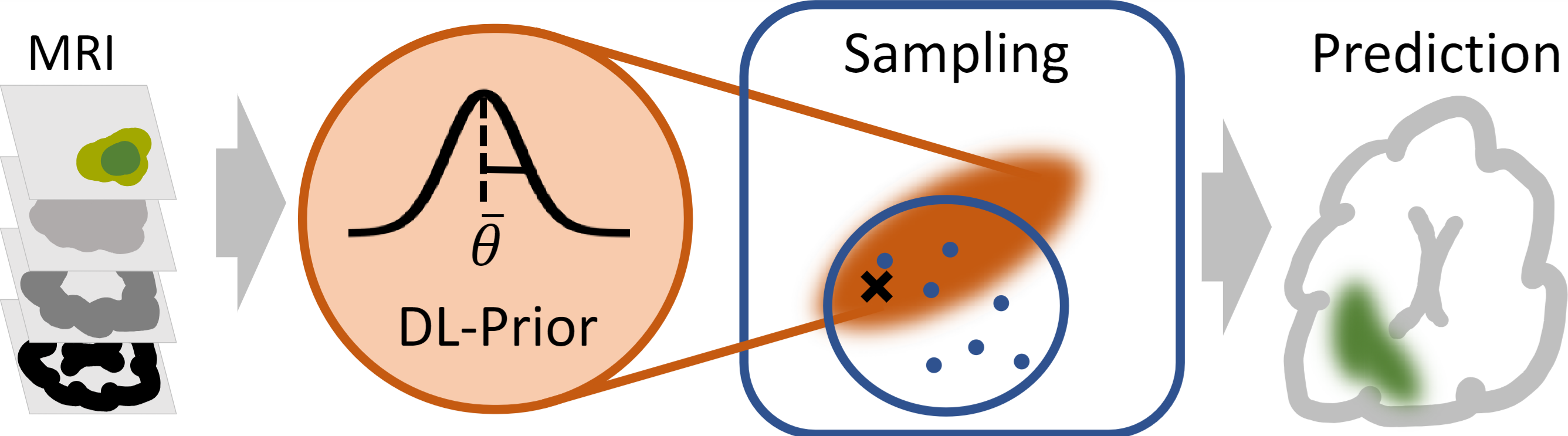 Biophysical modeling, particularly using partial differential equations (PDEs), offers significant potential to tailor disease treatment protocols to individual patients. However, the inverse problem-solving aspect of these models presents a substantial challenge, either due to the high computational requirements of model-based approaches or the limited robustness of deep learning (DL) methods. We propose a novel framework that leverages the unique strengths of both approaches in a synergistic manner. Our method incorporates a DL ensemble for initial parameter estimation, facilitating efficient downstream evolutionary sampling initialized with this DL-based prior.
Biophysical modeling, particularly using partial differential equations (PDEs), offers significant potential to tailor disease treatment protocols to individual patients. However, the inverse problem-solving aspect of these models presents a substantial challenge, either due to the high computational requirements of model-based approaches or the limited robustness of deep learning (DL) methods. We propose a novel framework that leverages the unique strengths of both approaches in a synergistic manner. Our method incorporates a DL ensemble for initial parameter estimation, facilitating efficient downstream evolutionary sampling initialized with this DL-based prior.
| [Paper] | [Code] |
VariViT
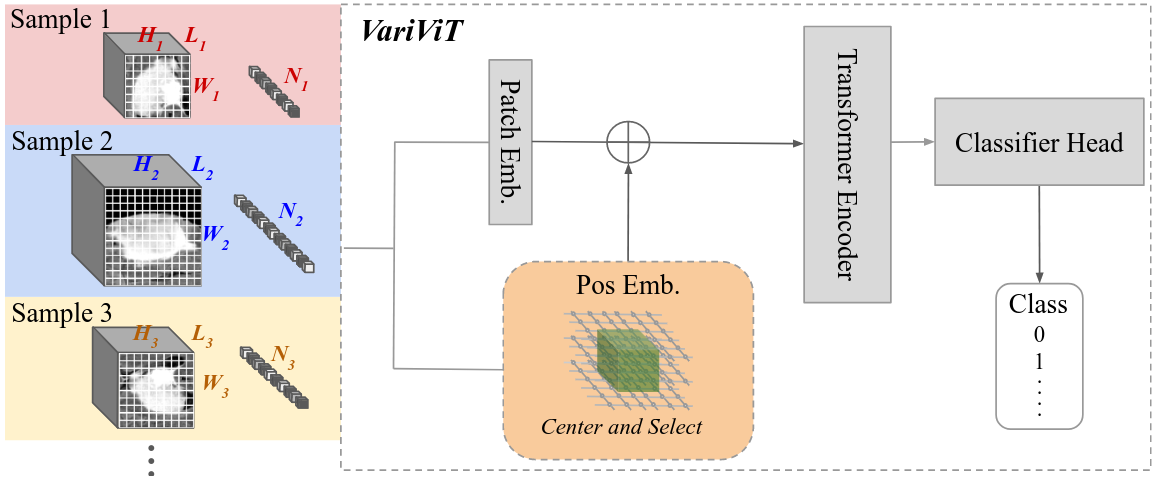 Vision Transformers (ViTs) have emerged as the state-of-the-art architecture for representation learning, leveraging self-attention mechanisms to excel across a range of tasks. ViTs split images into fixed-size patches, constraining them to a predefined size and requiring pre-processing steps such as resizing, padding, or cropping. This poses challenges in medical imaging, particularly for irregularly shaped structures such as tumors. To address this, we propose VariViT, an improved ViT model crafted to handle variable image sizes while maintaining a consistent patch size. VariViT employs a novel positional embedding resizing scheme for a variable number of patches. We also implement a new batching strategy within VariViT to reduce computational complexity, resulting in faster training and inference times.
Vision Transformers (ViTs) have emerged as the state-of-the-art architecture for representation learning, leveraging self-attention mechanisms to excel across a range of tasks. ViTs split images into fixed-size patches, constraining them to a predefined size and requiring pre-processing steps such as resizing, padding, or cropping. This poses challenges in medical imaging, particularly for irregularly shaped structures such as tumors. To address this, we propose VariViT, an improved ViT model crafted to handle variable image sizes while maintaining a consistent patch size. VariViT employs a novel positional embedding resizing scheme for a variable number of patches. We also implement a new batching strategy within VariViT to reduce computational complexity, resulting in faster training and inference times.
| [Paper] | [Code] |
LST-AI
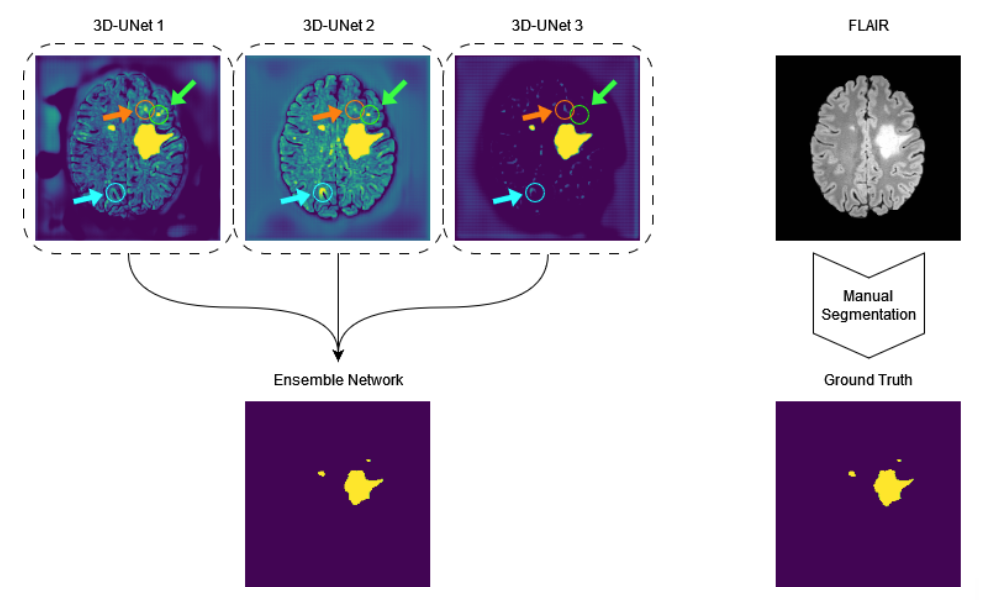 LST-AI is a publicly available UNet ensemble that segments inflammatory white matter MS lesions and classifies their location according to the 2017 McDonald criteria.
LST-AI is a publicly available UNet ensemble that segments inflammatory white matter MS lesions and classifies their location according to the 2017 McDonald criteria.
| [Paper] | [Code] |
INR for Multi-Contrast Super-Resolution
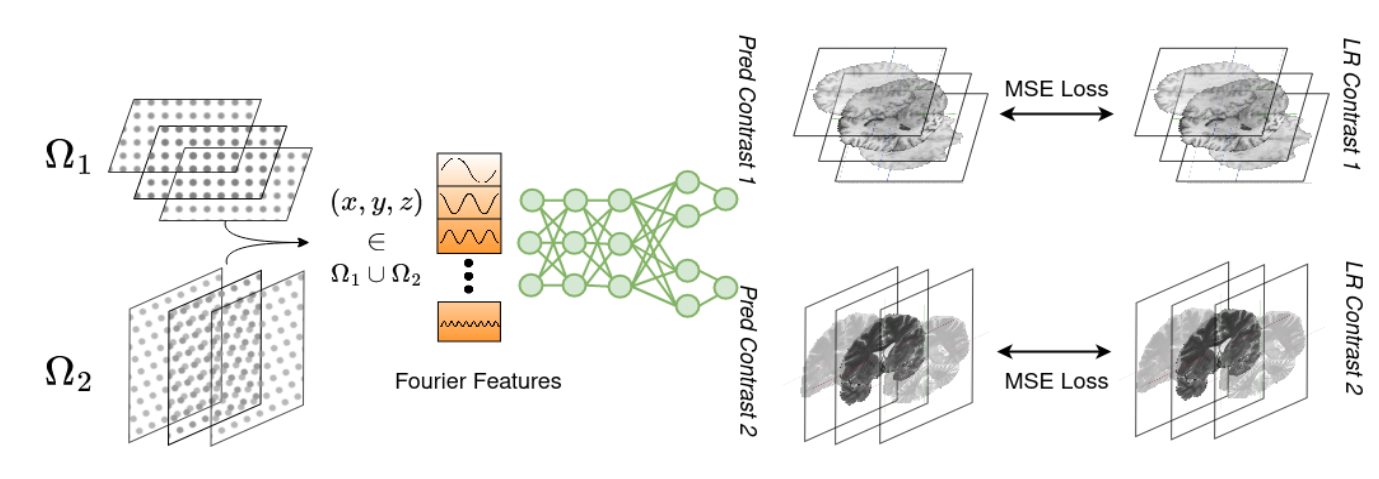 Combining different views of multi-contrast scans into high-resolution, isotropic 3D scans is challenging due to the limited availability of large training cohorts, which calls for a subject-specific framework. This work proposes a novel solution to this problem, leveraging Implicit Neural Representations (INR). Our proposed INR jointly learns two different contrasts of complementary views in a continuous spatial function and benefits from exchanging anatomical information between them.
Combining different views of multi-contrast scans into high-resolution, isotropic 3D scans is challenging due to the limited availability of large training cohorts, which calls for a subject-specific framework. This work proposes a novel solution to this problem, leveraging Implicit Neural Representations (INR). Our proposed INR jointly learns two different contrasts of complementary views in a continuous spatial function and benefits from exchanging anatomical information between them.
| [Paper] | [Code] |
GraphMS
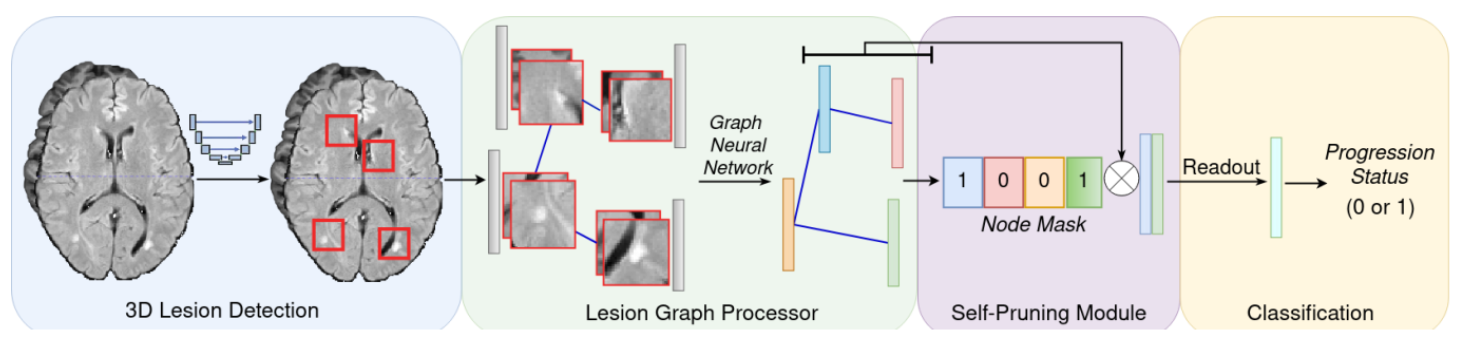 Our work represents the first attempt to use graph neural networks (GNNs) to aggregate lesion-based imaging biomarkers into a novel global representation. We propose a two-stage approach to predicting MS inflammatory disease activity. First, a 3D segmentation network detects lesions, and a self-supervised algorithm extracts their image features. Second, the detected lesions are used to build a patient graph. The lesions act as nodes in the graph and are initialized with image features extracted in the first stage. Finally, the lesions are connected based on their spatial proximity, and the inflammatory disease activity prediction is formulated as a graph classification task. Furthermore, we propose a self-pruning strategy to auto-select the most critical lesions for prediction.
Our work represents the first attempt to use graph neural networks (GNNs) to aggregate lesion-based imaging biomarkers into a novel global representation. We propose a two-stage approach to predicting MS inflammatory disease activity. First, a 3D segmentation network detects lesions, and a self-supervised algorithm extracts their image features. Second, the detected lesions are used to build a patient graph. The lesions act as nodes in the graph and are initialized with image features extracted in the first stage. Finally, the lesions are connected based on their spatial proximity, and the inflammatory disease activity prediction is formulated as a graph classification task. Furthermore, we propose a self-pruning strategy to auto-select the most critical lesions for prediction.
| [Paper] | [Code] |